Seed Funding Program 2023
PRIMA
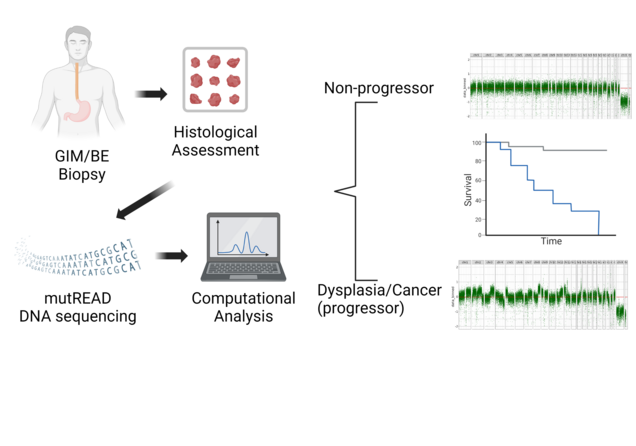
Development of biomarker predicting progression risk of Barrett’s Oesophagus
Coordinator

Sebastian Belle
II. Department of Medicine, UMM

Karol Nowicki-Osuch
DKFZ-Hector Junior Research Group Tumorigenesis and molecular cancer prevention
Detailed description
Personalized T cell therapies are both expensive and time consuming to develop and manufacture. For preparation of biologics, the Paul Ehrlich Institute (PEI, Federal Institute for Vaccines and Biomedicines) requires robust validation data to assess and issue market authorization of biomedicines for human use for new personalized approaches. In my previous work, I have shown that tumor-reactive T cells and their corresponding receptors TCR can be identified using “predicTCR”; a machine learning model trained on single cell RNA-seq and tumor reactivity data. However, the number of suitable datasets are often limited by the availability of tumor material that can be used for reactivity validation. By utilizing individualized patient-derived tumor organoids (IPTOs) developed in the division of Haikun Liu (DKFZ), we can routinely generate tumor organoids for reactivity validation, thereby increasing the amount of validated datasets for training while also demonstrating proof-of-concept data. Furthermore, the “Hamburger linear assembly”, developed in working group of Edward Green (DKFZ), reduces the time required to generate testable TCRs. Together, the Combined 3 Platform personalized Onco-immunotherapeutics (C-3PO) approach that I am developing will allow the rapid validation of patient-specific tumor-reactive TCR.